Science
Scientists Map Bacterial Kinases, Unveiling New Antibiotic Targets
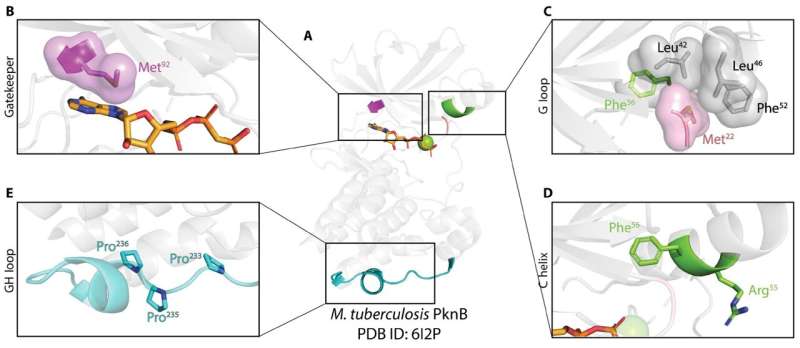
Researchers at the University of Georgia have created a comprehensive atlas of bacterial kinases, identifying potential targets for new antibiotic treatments. This extensive compendium focuses on serine-threonine kinases, enzymes critical for regulating cell growth and pathogenicity in various bacterial species. By analyzing nearly 26,000 strains of bacteria, the team aims to advance the fight against antibiotic resistance.
Dr. Brady O’Boyle, the lead author of the study, emphasizes that bacterial serine-threonine kinases influence numerous cellular processes, including virulence. The compendium allows researchers to explore how these kinases differ from their eukaryotic counterparts, providing insights that could lead to innovative methods for combating bacterial infections.
The study highlights that the number of serine-threonine kinases varies significantly across bacterial genomes, with some species, such as those in the Actinobacteria group, containing over 60 kinases. In contrast, the common bacterium Escherichia coli possesses only one. This variability presents exciting opportunities for targeted drug development.
The significance of serine-threonine kinases in humans has long been recognized, as these enzymes are involved in numerous diseases. For instance, medications like everolimus, an mTOR inhibitor, target serine-threonine kinases to manage conditions such as kidney cancer. By investigating the bacterial versions of these enzymes, researchers hope to find new strategies for antibiotic therapies that circumvent existing resistance mechanisms.
In constructing their atlas, the research team focused on Hanks-type serine-threonine protein kinases. These enzymes are essential in various biological processes, including metabolism and cell signaling. The phosphorylation process, in which a phosphate group is added to specific amino acids in a protein, is vital for regulating protein functions.
The findings, published in Science Signaling, detail the classification of over 300,000 bacterial serine-threonine kinase sequences sourced from the National Center for Biotechnology Information’s reference sequence database. This extensive analysis aids in understanding the evolutionary relationships between bacterial and eukaryotic kinases.
As researchers delve deeper into the compendium, they have identified novel pathways to target bacteria like Mycobacterium tuberculosis. Initial biochemical screenings suggest that inhibiting serine-threonine kinase activity could effectively control this pathogen, which poses significant global health challenges.
The team classified the kinases into 35 canonical families and seven pseudokinase families, based on shared similarities in their catalytic domains. This classification not only aids researchers in identifying specific bacterial kinases but also highlights the evolutionary differences between bacterial and eukaryotic kinases.
O’Boyle notes the potential for this atlas to inform the development of broad-spectrum antimicrobial drugs targeting bacterial serine-threonine kinases. The research underscores a crucial step in addressing the growing crisis of antibiotic resistance, offering a pathway to innovative therapeutic options.
As the study unfolds, the compendium serves as a vital resource for scientists exploring bacterial signaling and the development of selective kinase inhibitors. With the ongoing threat of drug-resistant infections, advancements in this field could have far-reaching implications for global health.
-
 Technology5 months ago
Technology5 months agoDiscover the Top 10 Calorie Counting Apps of 2025
-

 Technology3 weeks ago
Technology3 weeks agoOpenAI to Implement Age Verification for ChatGPT by December 2025
-

 Health3 months ago
Health3 months agoBella Hadid Shares Health Update After Treatment for Lyme Disease
-

 Health4 months ago
Health4 months agoAnalysts Project Stronger Growth for Apple’s iPhone 17 Lineup
-

 Health4 months ago
Health4 months agoErin Bates Shares Recovery Update Following Sepsis Complications
-

 Technology5 months ago
Technology5 months agoDiscover How to Reverse Image Search Using ChatGPT Effortlessly
-

 Technology3 months ago
Technology3 months agoElectric Moto Influencer Surronster Arrested in Tijuana
-

 Technology5 months ago
Technology5 months agoMeta Initiates $60B AI Data Center Expansion, Starting in Ohio
-

 Technology2 months ago
Technology2 months agoDiscover 2025’s Top GPUs for Exceptional 4K Gaming Performance
-
 Technology5 months ago
Technology5 months agoRecovering a Suspended TikTok Account: A Step-by-Step Guide
-

 Health5 months ago
Health5 months agoTested: Rab Firewall Mountain Jacket Survives Harsh Conditions
-

 Lifestyle5 months ago
Lifestyle5 months agoBelton Family Reunites After Daughter Survives Hill Country Floods















