Science
AI Discovers New Antibiotics from Ancient Archaea Microbes
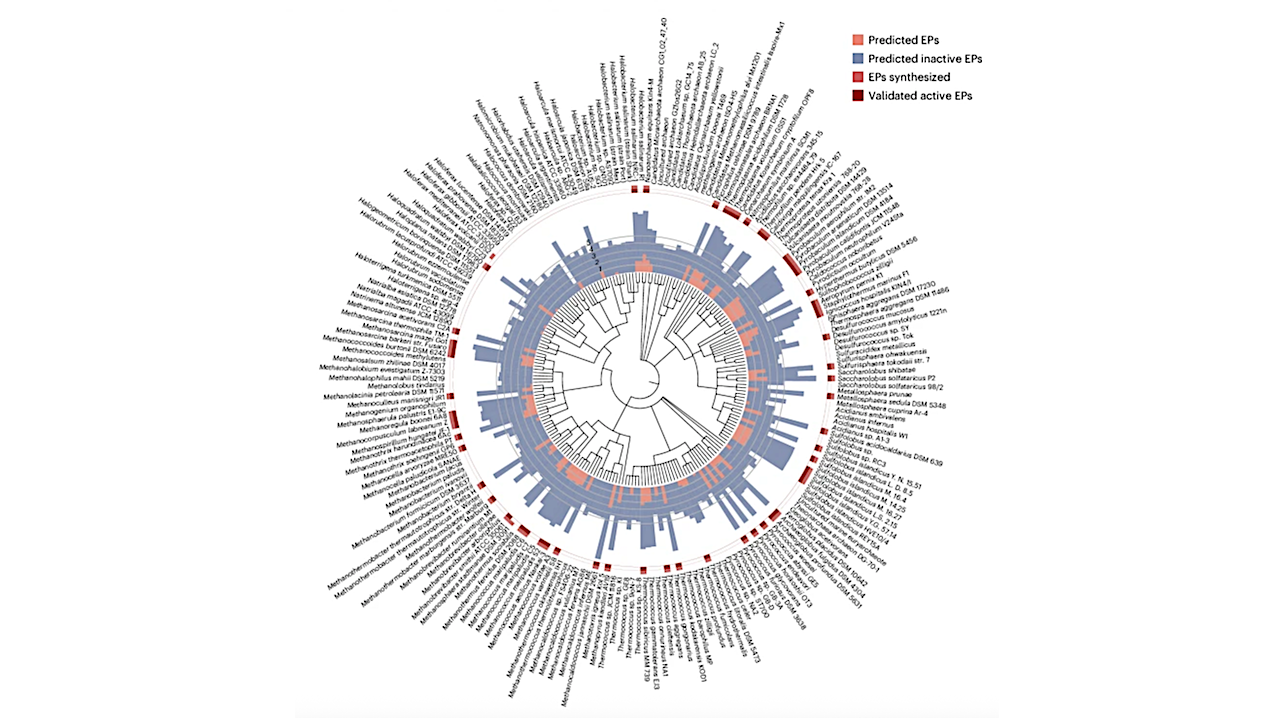
Research from the University of Pennsylvania has unveiled promising new antibiotic compounds derived from ancient microbes known as Archaea. These microbes, which have thrived in extreme environments for billions of years, may provide critical solutions to combat the rising threat of antibiotic resistance. The study, published in Nature Microbiology, utilized artificial intelligence to identify previously unknown compounds that could lead to the next generation of antibiotics.
According to César de la Fuente, Presidential Associate Professor at the university, traditional antibiotic discovery methods have largely focused on fungi, bacteria, and animals. His lab has previously employed AI to uncover antibiotic candidates from various unconventional sources, including the DNA of extinct species and animal venoms. Now, they are applying these advanced tools to analyze the proteins of hundreds of Archaea species.
Exploring the Unique World of Archaea
Archaea represent a distinct branch of life that differs fundamentally from both bacteria and eukaryotes, which encompass plants and animals. These microorganisms exhibit unique genetic and biochemical characteristics that enable them to survive in some of the planet’s most hostile environments, such as deep-sea vents and geothermal springs. This resilience suggests that Archaea may possess unique biochemical defenses that could inspire novel antibiotics.
Research associate Marcelo Torres, co-first author of the study, emphasized the potential of Archaea to provide insights into microbial competition. “If they’ve survived for billions of years under extreme conditions,” he noted, “maybe they’ve developed unique ways to fight off microbial competitors.”
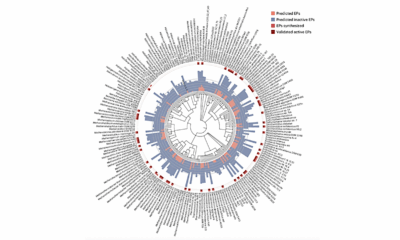
To identify antibiotic compounds within Archaea, the research team harnessed an updated version of their AI tool, APEX. Originally designed to find antibiotic candidates in ancient biological data, APEX was retrained with thousands of additional peptide sequences and information about pathogenic bacteria. This allowed the tool to predict which peptides in Archaea might inhibit bacterial growth.
After scanning 233 species of Archaea, the researchers identified over 12,000 antibiotic candidates, which they named “archaeasins.” Chemical analysis indicated that these molecules differ significantly from existing antimicrobial peptides, particularly in their electric charge distribution.
Promising Results Against Drug-Resistant Bacteria
The team proceeded to test 80 archaeasins against various disease-causing bacteria. Remarkably, 93% of these compounds demonstrated antimicrobial activity against at least one bacterium. The researchers selected three archaeasins for further testing in animal models, observing that all three halted the spread of a common drug-resistant bacterium found in hospitals just four days after a single dose. One of these compounds showed efficacy comparable to polymyxin B, a last-resort antibiotic for treating resistant infections.
“This research shows that there are potentially many antibiotics waiting to be discovered in Archaea,” de la Fuente stated. As antibiotic resistance becomes an increasingly critical issue, the search for new compounds in unconventional sources is paramount.
The study received funding from various organizations, including the Procter & Gamble Company and United Therapeutics, and was supported by grants from the National Institutes of Health and the Langer Prize.
Future research will focus on enhancing APEX to predict antibiotic candidates based on their structural properties, which could improve accuracy in identifying promising compounds. The team also aims to assess the long-term efficacy and safety of archaeasins, with the ultimate goal of advancing these compounds into human clinical trials.
“This is only the beginning,” de la Fuente remarked. “Archaea is one of the oldest forms of life, and clearly has much to teach us about how to outsmart the pathogens we face today.” As antibiotic resistance continues to escalate, the exploration of ancient microbial life may hold the key to developing effective new therapies.
-
 Technology4 weeks ago
Technology4 weeks agoDiscover the Top 10 Calorie Counting Apps of 2025
-

 Lifestyle1 month ago
Lifestyle1 month agoBelton Family Reunites After Daughter Survives Hill Country Floods
-

 Education1 month ago
Education1 month agoWinter Park School’s Grade Drops to C, Parents Express Concerns
-

 Technology2 weeks ago
Technology2 weeks agoDiscover How to Reverse Image Search Using ChatGPT Effortlessly
-

 Technology3 weeks ago
Technology3 weeks agoHarmonic Launches AI Chatbot App to Transform Mathematical Reasoning
-

 Technology1 month ago
Technology1 month agoMeta Initiates $60B AI Data Center Expansion, Starting in Ohio
-

 Lifestyle1 month ago
Lifestyle1 month agoNew Restaurants Transform Minneapolis Dining Scene with Music and Flavor
-

 Technology1 month ago
Technology1 month agoByteDance Ventures into Mixed Reality with New Headset Development
-

 Technology4 weeks ago
Technology4 weeks agoMathieu van der Poel Withdraws from Tour de France Due to Pneumonia
-
 Technology1 month ago
Technology1 month agoRecovering a Suspended TikTok Account: A Step-by-Step Guide
-

 Technology1 month ago
Technology1 month agoGlobal Market for Air Quality Technologies to Hit $419 Billion by 2033
-

 Health1 month ago
Health1 month agoSudden Vision Loss: Warning Signs of Stroke and Dietary Solutions